The Role of Crystallography-Grade Proteins in Advancing Structure-Based Drug Discovery
__548355399.png)
The Role of Crystallography-Grade Proteins in Advancing Structure-Based Drug Discovery
KRAS mutations have been a significant focus of cancer research for decades. These mutations affect a member of the RAS family of GTPases, which play a critical role in regulating cell division. The discovery of KRAS mutations dates back to the early 1980s when researchers first identified RAS oncogenes in human cancers. Since then, KRAS mutations have been strongly associated with poor prognosis in several cancers, including pancreatic, lung, and colorectal cancers (Cox et al., 2014; Stephen et al., 2014). Historically, KRAS mutants were labeled “undruggable” due to their challenging molecular properties, including the lack of well-defined binding pockets and their high affinity for GTP/GDP, which made targeting them with small molecules extremely difficult. However, structure-based drug design (SBDD) has transformed this landscape.
What is Structure-Based Drug Design (SBDD)?
Structure-Based Drug Design (SBDD) is a method in drug discovery that uses the 3D structure of a biological target, such as a protein, to design potential drug candidates. The process typically involves:
Determining the Structure: The target's structure is determined using techniques like X-ray crystallography or NMR spectroscopy.
Identifying Binding Sites: Researchers analyze the protein to locate binding pockets where a drug could interact effectively.
Designing Compounds: Computational modeling and molecular docking are used to design and optimize small molecules that fit into the binding site.
Validating Hits: The designed compounds are synthesized and tested experimentally for efficacy and binding affinity.
In the context of KRAS mutations, SBDD enabled the discovery of inhibitors targeting previously undruggable sites by leveraging structural information about the mutant protein's binding pocket. This approach offers significant advantages, including higher precision in drug design, reduced time and cost in lead optimization, and an increased likelihood of success by focusing on well-characterized binding sites. By providing a detailed understanding of the molecular interactions between the target and potential drug candidates, SBDD facilitates the rational design of compounds with improved efficacy and safety profiles.
In 2013, researchers discovered a novel allosteric site on the KRAS G12C mutant, leading to the development of effective inhibitors using x-ray crystallography.
Details of the Novel Allosteric Site on KRAS G12C
The "novel allosteric site" refers to a previously unexplored region on the KRAS G12C protein where small molecules can bind and inhibit its activity. An allosteric site is distinct from the active site of the protein. When a molecule binds to this site, it induces a conformational change that disrupts the protein’s function. This site on KRAS G12C enabled the development of covalent inhibitors, which bind selectively to the mutant form of KRAS without affecting the wild-type KRAS protein. The identification of this site was critical in overcoming the long-standing challenge of drugging KRAS mutants, which previously lacked obvious druggable pockets and displayed high affinity for their natural ligands, GTP and GDP. (Ostrem et al., 2013, 2016). This pivotal discovery made KRAS G12C mutants druggable, sparking new hope for researchers and patients. Further advancements were made in 2019 when Kessler et al. identified a non-specific KRAS inhibitor targeting a pocket between switch I and II on both active and inactive KRAS (Kessler et al., 2019).
Aurora Biolabs has played a critical role in the development of KRAS inhibitors through its expertise in SBDD. Notably, Aurora Biolabs' scientists were instrumental in the production, screening, and co-crystallization of complexes that led to the identification of a KRAS G12D inhibitor (Wang et al., 2022, Mirati Therapeutics). This mutation, KRAS G12D, is the most prevalent among KRAS mutations associated with pancreatic cancer.
The success of the Mirati Therapeutics project highlights Aurora Biolabs' comprehensive capabilities, offering a one-stop solution for drug discovery—from the production of crystallography-grade proteins to assay development, screening, co-crystallization, and data analysis. To learn more, visit our Services page or Contact Us for tailored solutions.
References
Cox, A. D., et al. (2014). Drugging the undruggable RAS: Mission possible? Nature Reviews Drug Discovery, 13(11), 828-851.
Stephen, A. G., et al. (2014). Dragging Ras Back in the Ring. Cancer Cell 2014
Ostrem, J. M., et al. (2013). K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature, 503(7477), 548-551.
Jonathan M.L. Ostrem, and Kevan M. Shokat, et al. (2016). Targeting KRAS G12C with Covalent Inhibitors. Annual Review of Cancer Biology 6, pp. 49 - 64 (2022)
Kessler, D., et al. (2019). Drugging an undruggable pocket on KRAS. Proceedings of the National Academy of Sciences, 116(32), 15823-15829.
Wang, X., et al. (2022). Discovery of KRAS G12D inhibitors for the treatment of pancreatic cancer. Journal of Medicinal Chemistry, 65(5), 2456-2470.
Available Products
Crystallography-grade WT and mutant KRAS proteins and Assay Kits for your drug discovery campaigns:
| Item Number | Mutations | Tag | Load |
|---|---|---|---|
| 5727-4121G | Kras Wild Type (WT) | GST-Tag | |
| 5727-WTG-G | Kras Wild Type (WT) | GST-Tag | GDP Loaded |
| 5727-WTG-GP | Kras Wild Type (WT) | GST-Tag | GppNHp loaded |
| 5727-4122H | Kras G12C (His-tag) | His-Tag | |
| 5727-4122G | Kras G12C (GST-tag) | GST-Tag | |
| 5727-4122G-G | Kras G12C (GST-tag) | GST-Tag | GDP Loaded |
| 5727-4122G-GP | Kras G12C (GST-tag) | GST-Tag | GppNHp Loaded |
| 5727-4123G | Kras G12D (GST-tag) | GST-Tag | |
| 5727-4123G-G | Kras G12D (GST-tag) | GST-Tag | GDP Loaded |
| 5727-4123G-GP | Kras G12D (GST-tag) | GST-Tag | GppNHp Loaded |
| 5727-4127G | Kras G12R (GST-tag) | GST-Tag | |
| 5727-4127G-G | Kras G12R (GST-tag) | GST-Tag | GDP Loaded |
| 5727-4127G-GP | Kras G12R (GST-tag) | GST-Tag | GppNHp Loaded |
| 5727-4128G | Kras G12V (GST-tag) | GST-Tag | |
| 5727-4128G-G | Kras G12V (GST-tag) | GST-Tag | GDP Loaded |
| 5727-4128G-GP | Kras G12V (GST-tag) | GST-Tag | GppNHp Loaded |
| 5727-4133G | Kras G13D (GST-tag) | GST-Tag | |
| 5727-4133G-G | Kras G13D (GST-tag) | GST-Tag | GDP Loaded |
| 5727-4133G-GP | Kras G13D (GST-tag) | GST-Tag | GppNHp Loaded |
KRAS Assay Kits
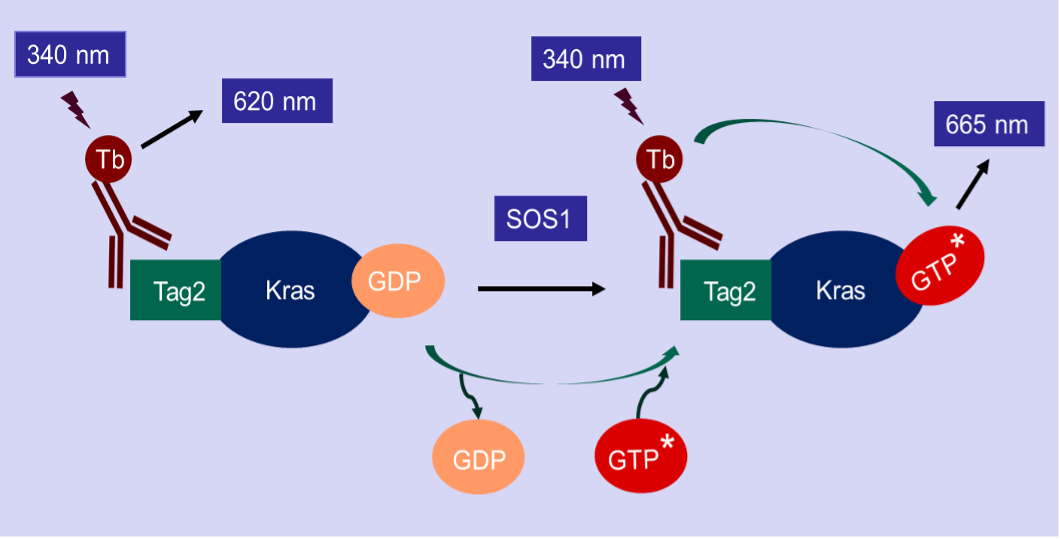
The Kras nucleotide exchange assay is a TR-FRET based assay. The assay kit is designed to detect the GTP binding status of Kras in the presence of SOS1, the most-studied guanine nucleotide exchange factor (GEF) of Kras. The Tag2 Kras in this assay kit is recognized by a Terbium-labeled anti-Tag2 antibody (HTRF donor). If Kras binds to a fluorescence-labeled GTP (HTRF acceptor), the donor and the acceptor will be brought in close proximity. Excitation of Terbium (340 nm) generates fluorescence resonance energy transfer (FRET) to the fluorescence-labeled GTP acceptor, which consequently fluoresces at 665 nm (figure below). Thus, GTP binding to Kras can be quantitively measured by calculation of the fluorescent ratio of 665 nm/620 nm.
| Item Number | Application |
|---|---|
| 5727-4121NK | Kras WT Nucleotide Exchange Assay Kit |
| 5727-4122NK | Kras G12C Nucleotide Exchange Assay Kit |
| 5727-4123NK | Kras G12D Nucleotide Exchange Assay Kit |
| 5727-4127NK | Kras G12R Nucleotide Exchange Assay Kit |
| 5727-4128NK | Kras G12V Nucleotide Exchange Assay Kit |
The Kras-cRAF binding assay kit is a TR-FRET based assay, which is designed to detect the binding status between Kras and cRAF. Tag2-Kras in this assay kit is loaded with GppNHp, which represents the activated Kras. The Ras binding domain (RBD) of cRAF has a Tag1 at N-terminus. A Terbium-labeled anti-Tag2 antibody binding to the Tag2-Kras serves as a fluorescence donor (HTRF donor), activation of which results in fluorescence resonance energy transfer (FRET) if the Tag1-cRAF binds to Kras, since the binding brings Terbium on the anti-Tag2 antibody close to the fluorophore on the anti-Tag1 antibody (HTRF acceptor). Thus, the binding status can be quantitively measured by calculating the ratio of the emission fluorescence intensity of the acceptor (665 nm) and donor (620 nm). Blocking the Kras-cRAF binding will reduce the HTRF signal. The ratio of the emission fluorescence intensity of the acceptor (665 nm) and donor (620 nm).

| Item Number | Application |
|---|---|
| 5727-4121BK | Kras WT – cRAF Binding Assay Kit |
| 5727-4122BK | Kras G12C – cRAF Binding Assay Kit |
| 5727-4123BK | Kras G12D– cRAF Binding Assay Kit |
Conclusion
The development of effective KRAS inhibitors, once deemed impossible, underscores the transformative power of structure-based drug design (SBDD). By leveraging advanced techniques such as X-ray crystallography and molecular modeling, researchers have identified novel binding sites and created inhibitors targeting previously undruggable KRAS mutations. Aurora Biolabs has been at the forefront of these efforts, offering a comprehensive suite of services, from producing crystallography-grade proteins to assay development and co-crystallization. These advancements not only open new avenues for cancer treatment but also highlight the importance of collaborative efforts in drug discovery. For researchers and pharmaceutical companies aiming to accelerate their drug development pipelines, Aurora Biolabs provides a trusted and capable partner in tackling the challenges of precision medicine. Additionally, all products described above are now available at BioHippo, enabling immediate access to high-quality KRAS proteins and assay kits to support your research.
 Loading ....
Loading ....
